Project Description
The Sardinian Transcriptome Project
The interpretation of GWAS is extremely challenging since most candidate loci fall in non-coding regions and are difficult to interpret. Given the important role of regulatory variation in shaping phenotypic complexity, expression Quantitative Trait Loci (eQTLs) have been proposed as informative intermediate phenotypes between genomic sequence variation and human disease.
Contacts:
mauro.pala@irgb.cnr.it
mara.marongiu@irgb.cnr.it
Keywords:
NGS, Expression profiles, eQTL
RNAseq in leukocytes
We are profiling with RNAseq the leukocytes of sardinian individuals enrolled in the ProgeNIA/SardiNIA project.
We initially mapped a collection of more than 30,000 eQTLs in freshly isolated peripheral blood leukocytes derived from 606 SardiNIA volunteers and the results have been published in Pala et al., 2017 and our results provide insight into the effects of regulatory variants and their relationship to population history and individual genetic risk.
The main findings were:
- We identified 21,183 expression quantitative trait loci (eQTLs) and 6,768 splicing quantitative trait loci (sQTLs), including 619 new QTLs.
- We found multiple independent eQTLs and sQTLs for the same gene.
- We found that QTLs show larger effect sizes in Sardinia than in Europe.
- We identified high-frequency QTLs and found evidence of selection near genes involved in malarial resistance and increased multiple sclerosis risk, reflecting the epidemiological history of Sardinia.
- Using family relationships, we identified 809 segregating expression outliers (median z score of 2.97), averaging 13.3 genes per individual. Outlier genes were enriched for proximal rare variants, providing a new approach to study large-effect regulatory variants and their relevance to traits.
eQTLs and sQTLs can be downloaded here
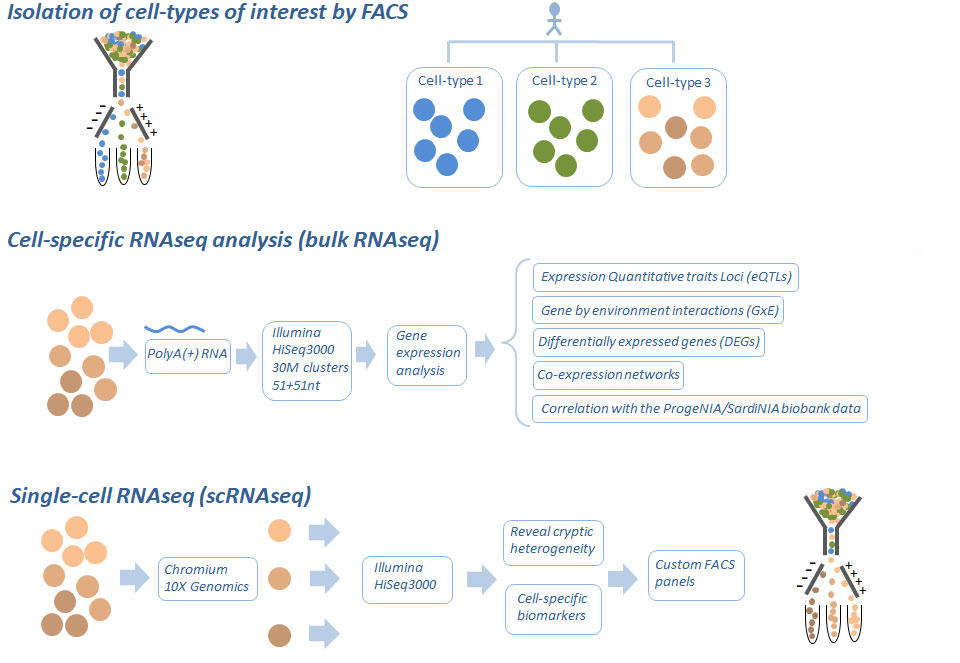
RNAseq in specific cell-types down to single-cells
We are aimed to identify and quantify gene expression profiles and establish their genetic regulation in the critical cell types related to autoimmunity, age and environmental factors.
We use RNA sequencing to characterize the transcriptional profile in the specific cell-types and to map expression quantitative trait loci (eQTLs), identify differentially expressed genes (DEGs), coexpression networks, interactions of genes by environment (GxE) and by age (GxA), and determine the correlation between the transcriptome and the ProgeNIA/SardiNIA biobank data.
We also sub-characterize the immune cell populations of interest at a deeper level of resolution by single cell RNA sequencing, in order to resolve cryptic cellular heterogeneity, identify cells sub-populations that are the source of eQTL signals and determine biomarkers of the cell cell subpopulations that can be used to design custom antibody panels to FACS profile large sample sets at low cost.
Resources and tools
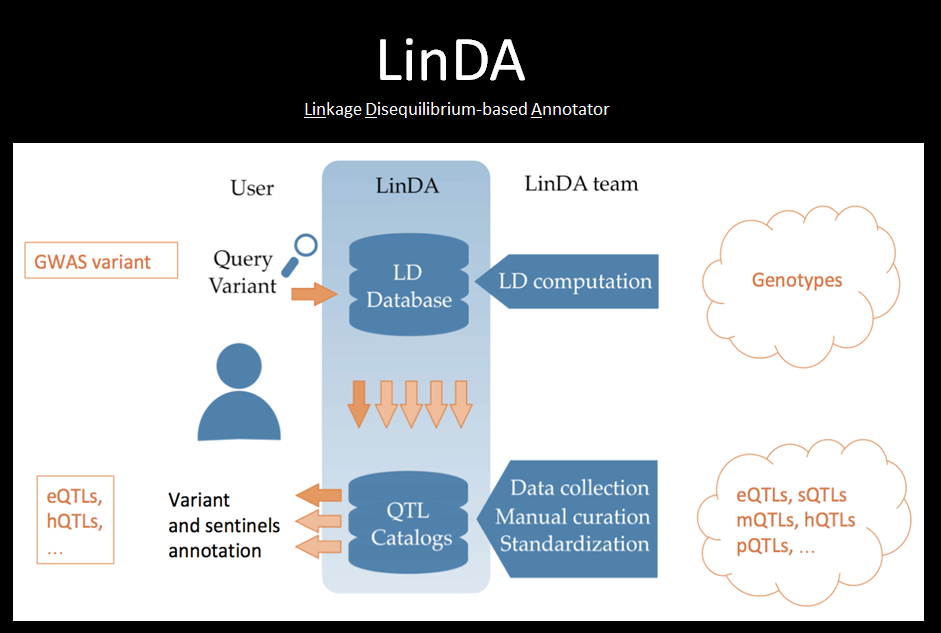
LinDA is a tool that, for a given variant, determines all the variants in Linkage Disequilibrium and whether they intersect with other association studies. A large manually curated collection of QTL and association studies can be easily interrogated and results are displayed with interactive plots and tables. LinDA offers also complementary tools, such as for computing the variant frequency and the pair-wise phasing, to facilitate the interpretation of the results.
The expression Qualitative Traits Loci (eQTLs) are genetic polymorphisms associated with changes in gene expression levels and they have been successfully used to interpret the molecular role of Genome Wide Association Studies (GWAS) variants. Up to date few eQTLs databases exist but they collect only a small portion of the available datasets. We are currently collecting and manually curating several studies. Here are the main links to the catalog